AI in Biotech
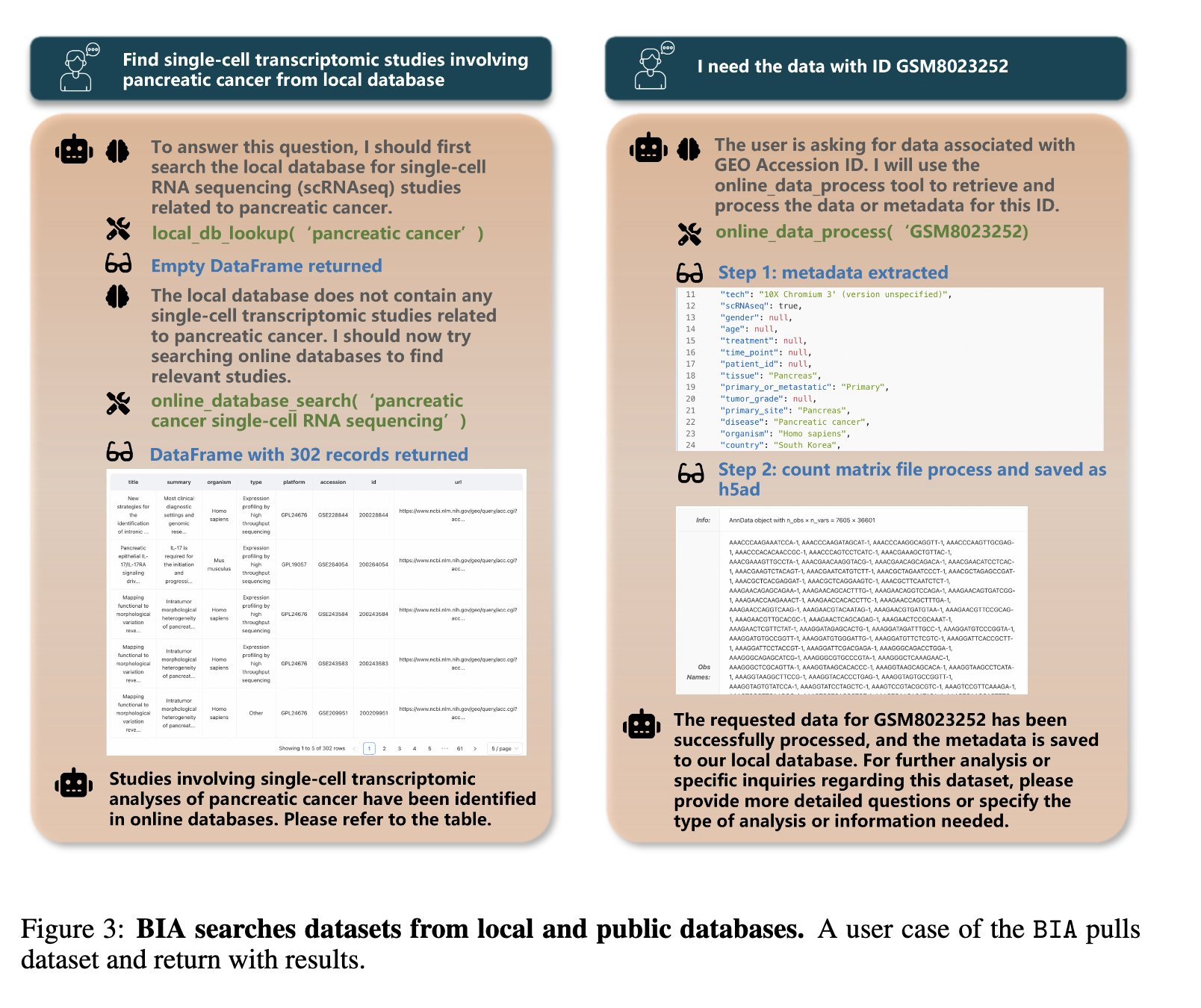
Bioinformatics Agent (BIA) is a Github repo “encompass extraction and processing of raw data and metadata, querying both locally deployed and public databases for information.”
Caduceus: Bi-Directional Equivariant Long-Range DNA Sequence Modelin
A sequence prediction model for DNA built on the Transformer competitor Mamba. It is extremely efficient and powerful for a small model. #gene #genetics
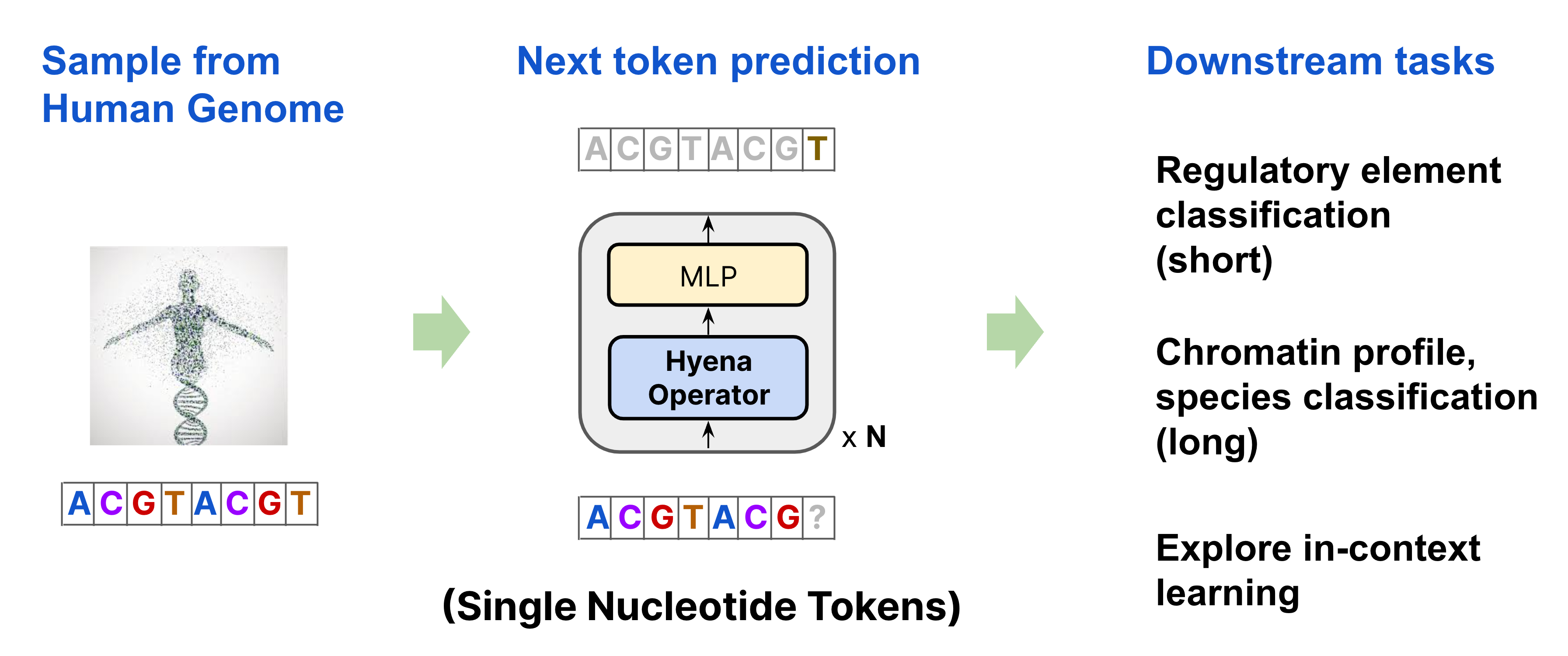
HyenaDNA is a Stanford-built model that uses transformers to predict qualities of the human genome
We’re excited to introduce HyenaDNA, a long-range genomic foundation model with context lengths of up to 1 million tokens at single nucleotide resolution!
Biological simulation
- Whole-body simulation of realistic fruit fly locomotion with deep reinforcement learning, HHMI Janelia, Google DeepMind. Introduces an anatomically detailed, biomechanical whole-body of a fruit fly for simulating realistic locomotion behaviors, such as flying and walking. Developed with the open source MuJoCo physics engines, the model includes sophisticated representations of the fly’s body parts, fluid forces during flight, and adhesion forces. Incorporating deep reinforcement learning, allows for the creation of neural network controllers that drive the simulated fly in complex trajectories and tasks based on sensory inputs, achieving high fidelity in locomotion simulation.
Proteins
ProtT3: Protein-to-Text Generation for Text-based Protein Understanding
combining Protein Language Models (PLMs) with traditional Language Models (LMs). ProtT3 integrates a PLM for processing amino acid sequences and a language model for generating high-quality textual descriptions using a cross-modal projector called Q-Former.
Google has released AlphaFold 3. Google DeepMind and Isomorphic Labs have developed the 3rd generation of AlphaFold, a powerful protein folding prediction model. Now it can predict 3D structure and interactions of all life’s molecules, like DNA, RNA and Ligands. AlphaFold 3 is 50% more accurate than previous generations. It correctly predicted the folded structure of the spike protein on Coronavirus OC43.
Access AlphaFold 3’s capabilities for free using the AlphaFold server.
Also see Jing, Berger, and Jaakkola (2024)
AlphaFold is used to predict the state of a protein after folding. By adding flow matching, which is invertible, you can dramatically improve modeling power on the entire landscape of proteins.
Sequence modeling and design from molecular to genome scale with Evo Sam Hammond thinks this is a really big deal.
Patrick Hsu: To aid our model design and scaling, we performed the first scaling laws analysis on DNA pretraining (to our knowledge) across leading architectures (Transformer++, Mamba, Hyena, and StripedHyena), training over 300 models from 6M to 1B parameters at increasing compute budgets
Evo is a protein language model, an RNA language model, and a regulatory DNA model 🤯
Evo can do prediction and generation across all 3 of these modalities. We show zero-shot function prediction across DNA, RNA, and protein modalities.
Samuel Hammond: SoTA zero-shot protein function prediction from a 7b parameter model. This alone justifies NVDA’s valuation. Every big pharma company is about to start pouring capex into training runs of their own. Text-to-organism is not far. If you doubted the Great Stagnation was over!
Challenges and Counterarguments
Quanta How AI Revolutionized Protein Science But Didn’t End It very detailed description of the history, challenges and solutions involved, culminating in AlphaFold, but notes some of the problems:
While AlphaFold2 is excellent at predicting the structures of small, simple proteins, it’s less accurate at predicting those containing multiple parts. It also can’t account for the protein’s environment or bonds with other molecules, which alter a protein’s shape in the wild. Sometimes a protein needs to be surrounded by certain ions, salts or metals to fold properly.
Importantly, AlphaFold2 can’t register point mutations, proteins that differ by only one amino acid.
AlphaFold3 and RoseTTAFold All-Atom — enable them to predict the structures of proteins bound to each other, DNA, RNA and other small molecules.
Problem: although AlphaFold mostly solves the protein folding problem, it can’t explain why it works. ***
The current science is based on a simple, straightforward model that assumes proteins generally fold only one way. But take A Holistic View of the Cell and it’s clear that life is far more complicated:
AlphaFold2, the computational model that predicts protein structures with an accuracy that matches or exceeds experimental methods, was trained mostly on protein structures solved with x-ray crystallography. But again, proteins in cells behave more like liquids than solids; they wiggle to-and-fro in a chaotic dance, and can adopt hundreds of different, distinct shapes.
If one reverses AlphaFold’s predictions, and instead makes the model generative, it tends to design proteins that are hyper-stable and rigid, much like the frozen proteins on which it was trained. This is part of the reason why it will be so difficult to design new functional proteins—AI models are not trained on a complete biological picture.
Cradle raised $47M for Protein engineering without the guesswork: “Design improved variants of your target protein sequence with just a few clicks — and some machine learning.”
Ex-Meta researchers founded EvolutionaryScale, raising $40M and valued at $200M to fast-track AI-driven protein structure predictions. With claims of outpacing Google’s AlphaFold in speed, the startup eyes breakthroughs in medicine and biotech
Here’s the document describing how it works: ESM3: Simulating 500 million years of evolution with a language model
.png)
Microsoft’s open source EvoDiff framework is a 640-million parameter model trained on data from all different species and functional classes of proteins.The data to train the model was sourced from the OpenFold data set for sequence alignments and UniRef50, a subset of data from UniProt, the database of protein sequence and functional information maintained by the UniProt consortium. Alamdari et al. (2023)

Google’s DeepMind AlphaMissence is a freely available AI catalog that has classified the potential effects of millions of missense genetic mutations, which could help establish the cause of diseases such as cystic fibrosis, sickle-cell anemia, and cancer.
The AlphaMissense resource from Google DeepMind categorized 89% of all 71 million possible missense variants.
Chemistry
ChemFlow: Navigating Chemical Space with Latent Flows. Enhance molecular science by efficiently navigating chemical space using deep generative models.
Navigating Chemical Space with Latent Flows by Guanghao Wei, Yining Huang, Chenru Duan, Yue Song, and Yuanqi Du.
Flows can uncover meaningful structures of latent spaces learned by generative models! We propose a unifying framework to characterize latent structures by flows/diffusions for optimization and traversal.

CRISPR editing
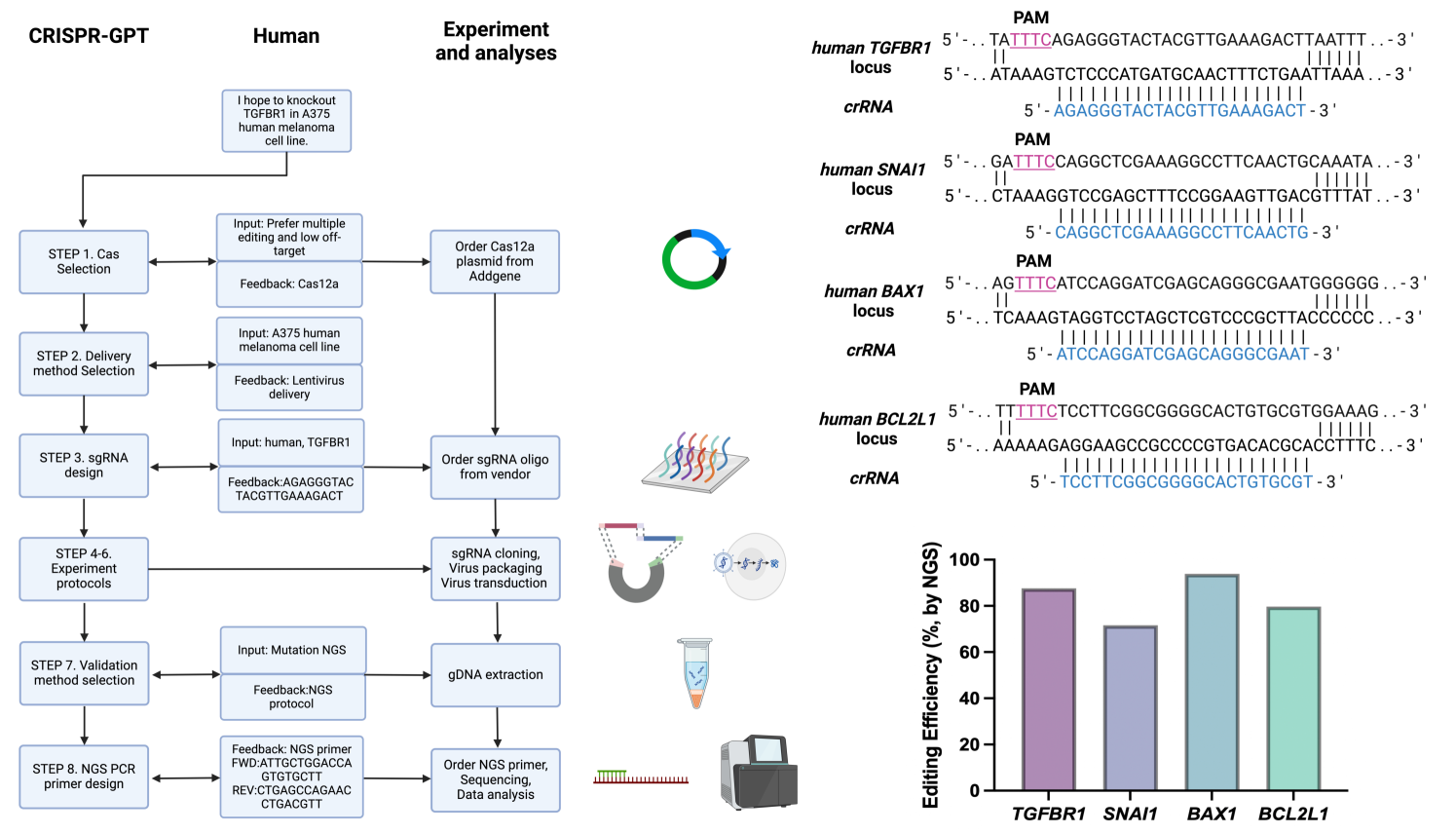
A Stanford-Princeton team that includes Russ Altman design a system intended to make easier the complex task of gene editing with CRISPR.
Because so many different tasks are involved, this approach uses agents that each handle different aspects of the problem.
Huang et al. (2024)
CRISPR-GPT leverages the reasoning ability of LLMs to facilitate the process of selecting CRISPR systems, designing guide RNAs, recommending cellular delivery methods, drafting protocols, and designing validation experiments to confirm editing outcomes
Synthetic biology
AI Accelerates Ability to Program Biology Like Software The Seattle-based synthetic biology startup Arzeda, co-founded by Alexandre Zanghellini, uses its Intelligent Protein Design Technology to design enzymes and protein sequences. The technology draws on generative AI in combination with a physics-based model.
Drugs
see @allthingsapx
Evo, a genetic foundation model from Arc Institute that learns across the fundamental languages of biology: DNA, RNA and proteins. Is DNA all you need?
Nathaniel Bennett, a computational biochemist at the University of Washington in Seattle developed Antibodies from scratch. They started with RFdiffusion, an AI tool that their team released last year2 that has helped to transform protein design. They modified it
based on a neural network similar to those used by image-generating AIs such as Midjourney and DALL·E. The team fine-tuned the network by training it on thousands of experimentally determined structures of antibodies attached to their targets, as well as real-world examples of other antibody-like interactions.